All organisms are able to adjust important cellular processes to the metabolic state. This includes the precise coordination of the production of proteins and other biomolecules according to the nutrient availability. To this end, organisms use molecules that act as direct indicators for the cellular metabolic state.
One of these central metabolites is activated acetic acid, so-called acetyl coenzyme A (short: acetyl-CoA). A high concentration of acetyl-CoA directly indicates high metabolic activity and high availability of nutrients, while a low concentration reflects deprivation of nutrients. Earlier reports showed that acetyl-CoA is used for enzymatically-catalysed and non-enzymatic acetylation of biomolecules. In order to re-establish the non-acetylated state and avoid the permanent indication of a cellular state with high nutrient availability, the cells use enzymes, the so-called deacetylases.
Notably, these enzymes are capable of not only deacetylating proteins and other biomolecules but also removing other acylations. All of these acyl-group modifications act as direct indicators for the cellular metabolic state. Furthermore, they can also reveal additional functional roles such as membrane targeting of proteins mediated by lysine acylation and deacylation. While this class of Zn2+-dependent classical deacylases has been studied extensively in eukaryotes such as mammals and plants, these enzymes had previously not been systematically explored in bacteria. This gap of knowledge has now been closed by this study.
“We have been able to elucidate a total of twelve crystal structures representing the five main groups of these bacterial enzymes. The concomitant functional characterisation of the activity and inhibition of these enzymes allowed us to gain a comprehensive overview of the structure and function of these bacterial enzymes,” explains PhD student and first author of this study Leonie Graf. The research group leader Prof. Michael Lammers added: “We did not expect to be able to identify so many uncharacterised enzymes of this class in bacteria. This project will serve as a basis for several follow-up projects on the physiological processes these enzymes are involved in and on the possibility of therapeutically controlling them. These comprehensive insights into this enzyme class were only made possible by the excellent collaboration with the research groups of Prof. Kay Hofmann from the University of Cologne, Prof. Richard Strugnell from the University of Melbourne, and Prof. Christian Olsen from the University of Copenhagen.”
Contributing groups:
Prof. Michael Lammers, University of Greifswald, Institute of Biochemistry
Prof. Kay Hofmann, University of Cologne
Prof. Christian A. Olsen, University of Copenhagen
Prof. Richard A. Strugnell, University of Melbourne
Further information
Research group of Prof. Dr. Michael Lammers
Article in the scientific journal Nature Communications (DOI 10.1038/s41467-024-53903-0)
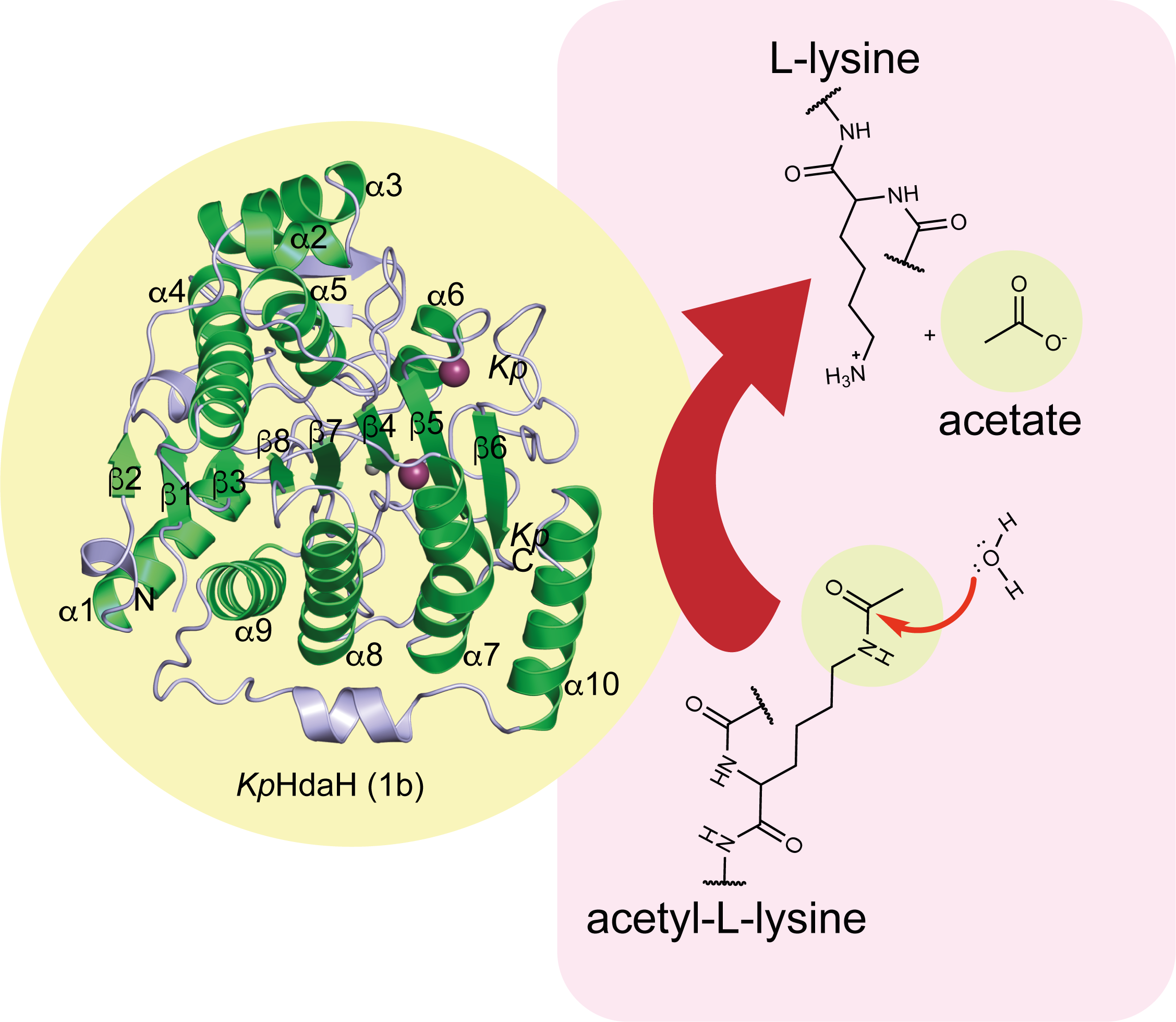
Illustration: Many bacterial species encode enzymes capable of deacytylating amino acids or other molecules such as the displayed enzyme HdaH from Klebsiella pneumoniae. © Michael Lammers
Contact at the University of Greifswald:
Prof. Dr. Michael Lammers
Institute of Biochemistry
Felix-Hausdorff-Straße 4
17489 Greifswald Tel.: +49 3834 420 4356
michael.lammersuni-greifswaldde